GitHub - tgac-vumc/ACE: Absolute Copy Number Estimation using low-coverage whole genome sequencing data
By A Mystery Man Writer
Absolute Copy Number Estimation using low-coverage whole genome sequencing data - tgac-vumc/ACE
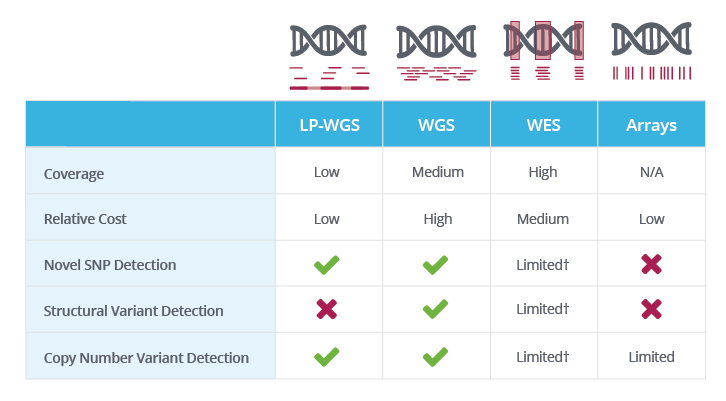
Low-Pass Whole Genome Sequencing
GitHub - yuchaojiang/CODEX2: Full-spectrum copy number variation detection by high-throughput DNA sequencing

Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data

GitHub - Nealelab/whole_genome_analysis_pipeline

Detection of Copy Number Variation using Shallow Whole Genome Sequencing Data to replace Array-Comparative Genomic Hybridization Analysis
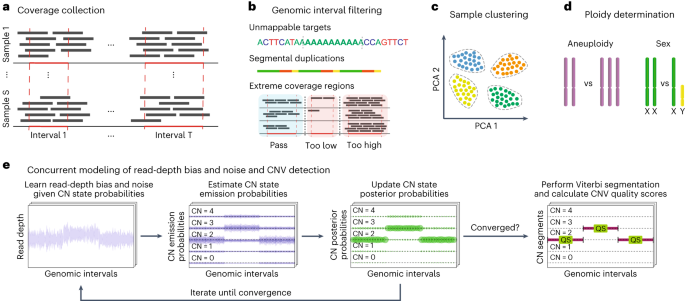
GATK-gCNV enables the discovery of rare copy number variants from exome sequencing data
Bamgineer: Introduction of simulated allele-specific copy number variants into exome and targeted sequence data sets

American Society for Clinical Pharmacology and Therapeutics - 2019 - Clinical Pharmacology & Therapeutics - Wiley Online Library

PDF) Low-Coverage Whole Genome Sequencing Using Laser Capture Microscopy with Combined Digital Droplet PCR: An Effective Tool to Study Copy Number and Kras Mutations in Early Lung Adenocarcinoma Development

CNAplot — Software for visual inspection of chromosomal copy number alteration in cancer using juxtaposed sequencing read depth ratios and variant allele frequencies - SoftwareX
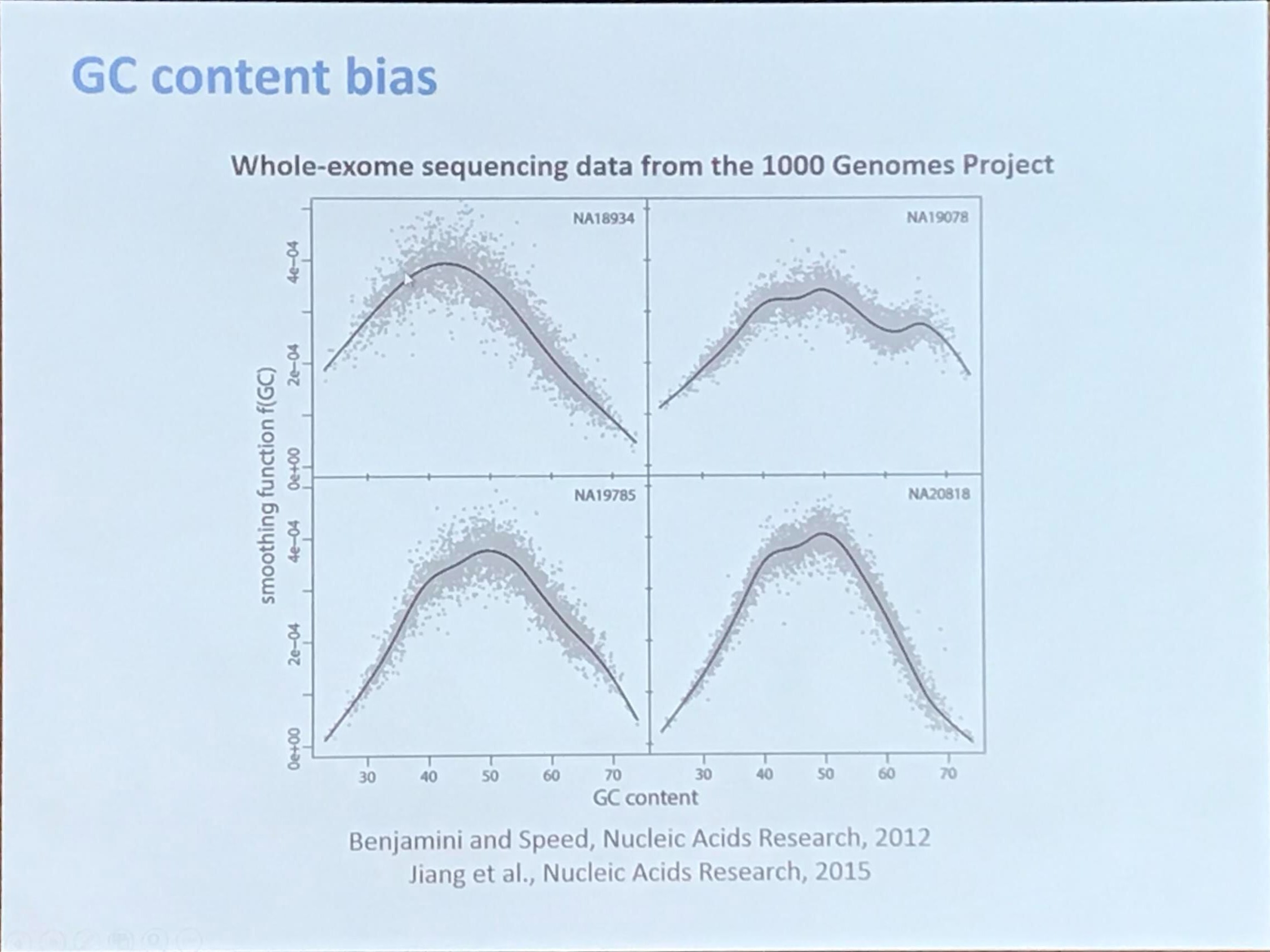
DNA copy number profiling: from bulk tissue to single cells

ClinSV: clinical grade structural and copy number variant detection from whole genome sequencing data, Genome Medicine

CODEX2: full-spectrum copy number variation detection by high-throughput DNA sequencing, Genome Biology
jpoell · GitHub
- Genotyping by low-coverage whole-genome sequencing in intercross pedigrees from outbred founders: a cost-efficient approach, Genetics Selection Evolution

- Capsule network-based approach for estimating grassland coverage using time series data from enhanced vegetation index - ScienceDirect

- JLPEA, Free Full-Text
- Site Selection for Land-Intensive Occupiers in Infill Areas — Chuck Berger

- Comparison of low-coverage and high-coverage sequencing reference for

- Mari Winsor Dead: Pilates Instructor to the Stars Was 70

- Nooyme Men's Thermal Underwear Set Long Johns for Men,Base Layer Blue NWT Size L

- Columbia Silver Ridge™ Utility Pants

- 1 Likes, 0 Comments - Elegant Sarees (@sareebazaar1) on Instagram: “🔥𝗦𝗮𝗹𝗲🔥𝗦𝗮𝗹𝗲 😍𝗢𝗿𝗱𝗲𝗿 𝗡𝗼𝘄😍@sareebazaar…

- Purple Brand Jeans Size 30 – St. John's Institute (Hua Ming)
