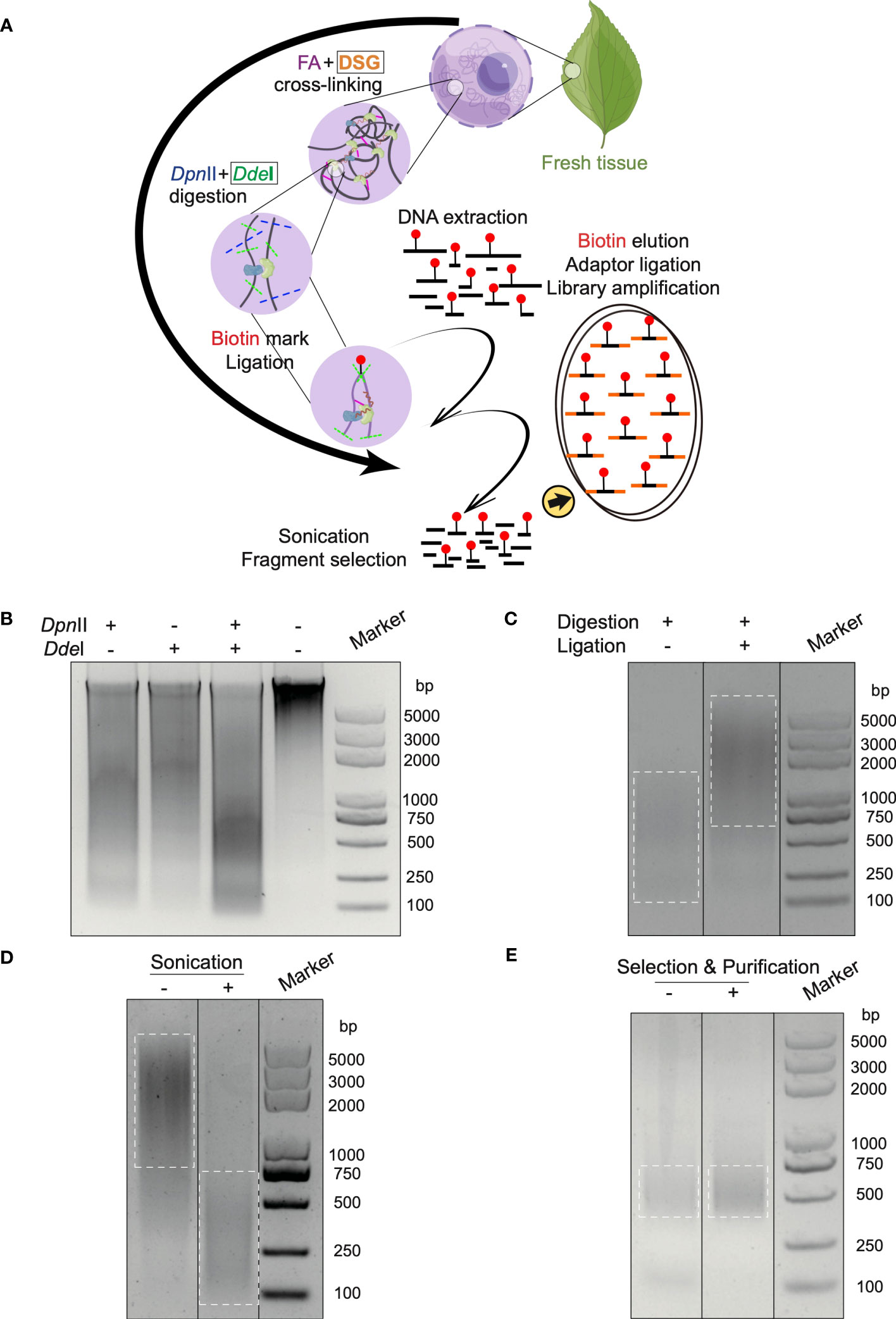
Fine fragmentation and DSG cross-linking improves loop detection a.

By A Mystery Man Writer

SPRITE: a genome-wide method for mapping higher-order 3D interactions in the nucleus using combinatorial split-and-pool barcoding
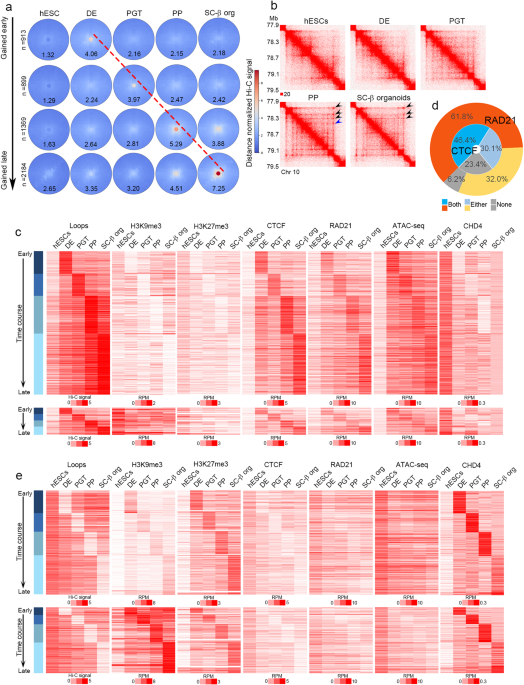
Regulation of CTCF loop formation during pancreatic cell differentiation

Krishna PARSI, Instructor, Doctor of Philosophy

Characterizing chromatin interactions of regulatory elements and nucleosome positions, using Hi-C, Micro-C, and promoter capture Micro-C, Epigenetics & Chromatin

A gas phase cleavage reaction of cross-linked peptides for protein complex topology studies by peptide fragment fingerprinting from large sequence database - ScienceDirect
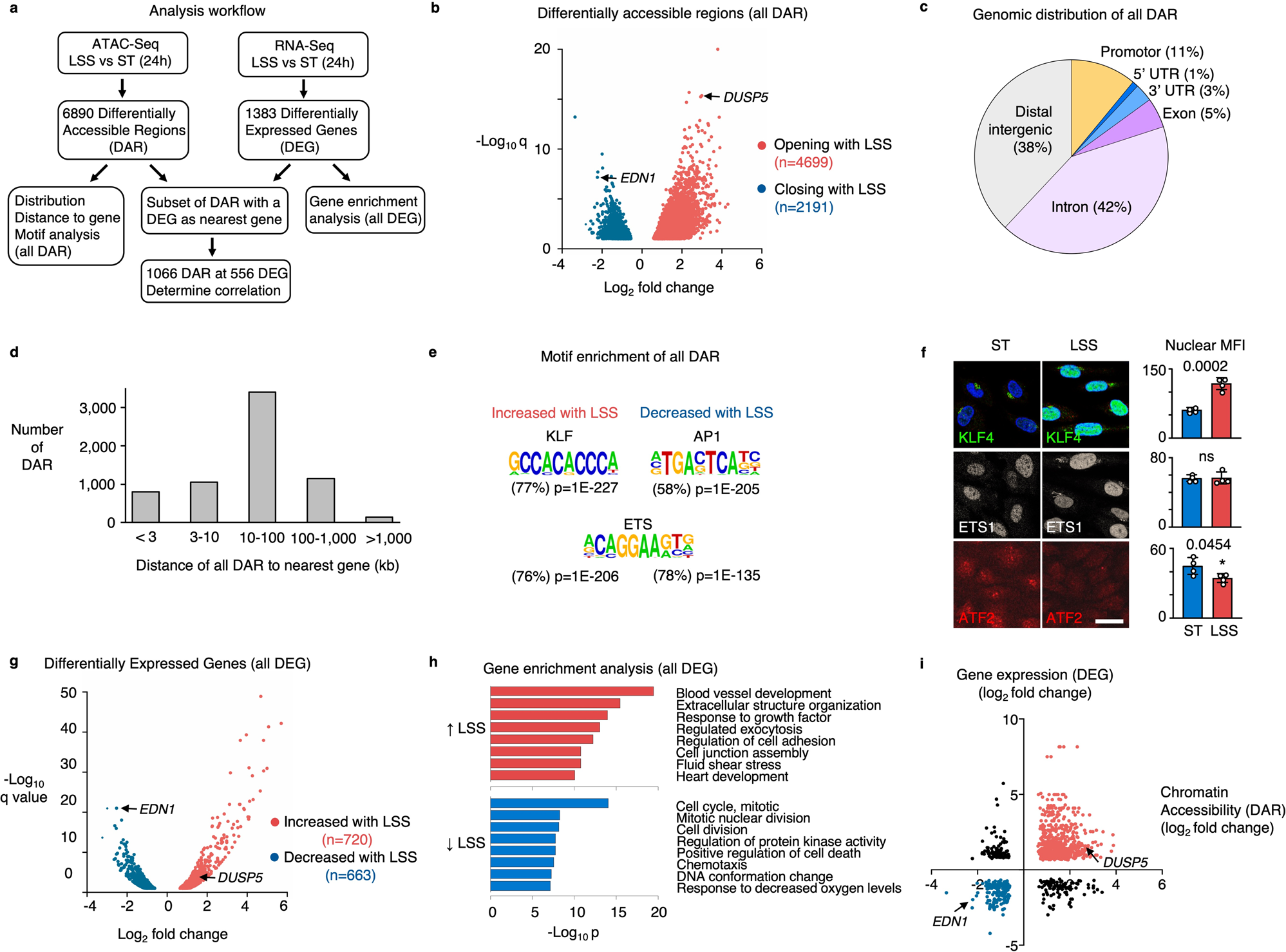
KLF4 recruits SWI/SNF to increase chromatin accessibility and reprogram the endothelial enhancer landscape under laminar shear stress

René Maehr's research works University of Massachusetts Medical

Principles and innovative technologies for decrypting noncoding RNAs: from discovery and functional prediction to clinical application, Journal of Hematology & Oncology

Insects, Free Full-Text

Johan GIBCUS University of Massachusetts Medical School
Frontiers An upgraded method of high-throughput chromosome conformation capture (Hi-C 3.0) in cotton (Gossypium spp.)

9: Chromatin loops are better detected in experiments with fine

Systematic evaluation of chromosome conformation capture assays

Regulation of CTCF loop formation during pancreatic cell differentiation

Job Dekker's research works Howard Hughes Medical Institute









