In silico prediction of immune-escaping hot spots for future COVID
By A Mystery Man Writer
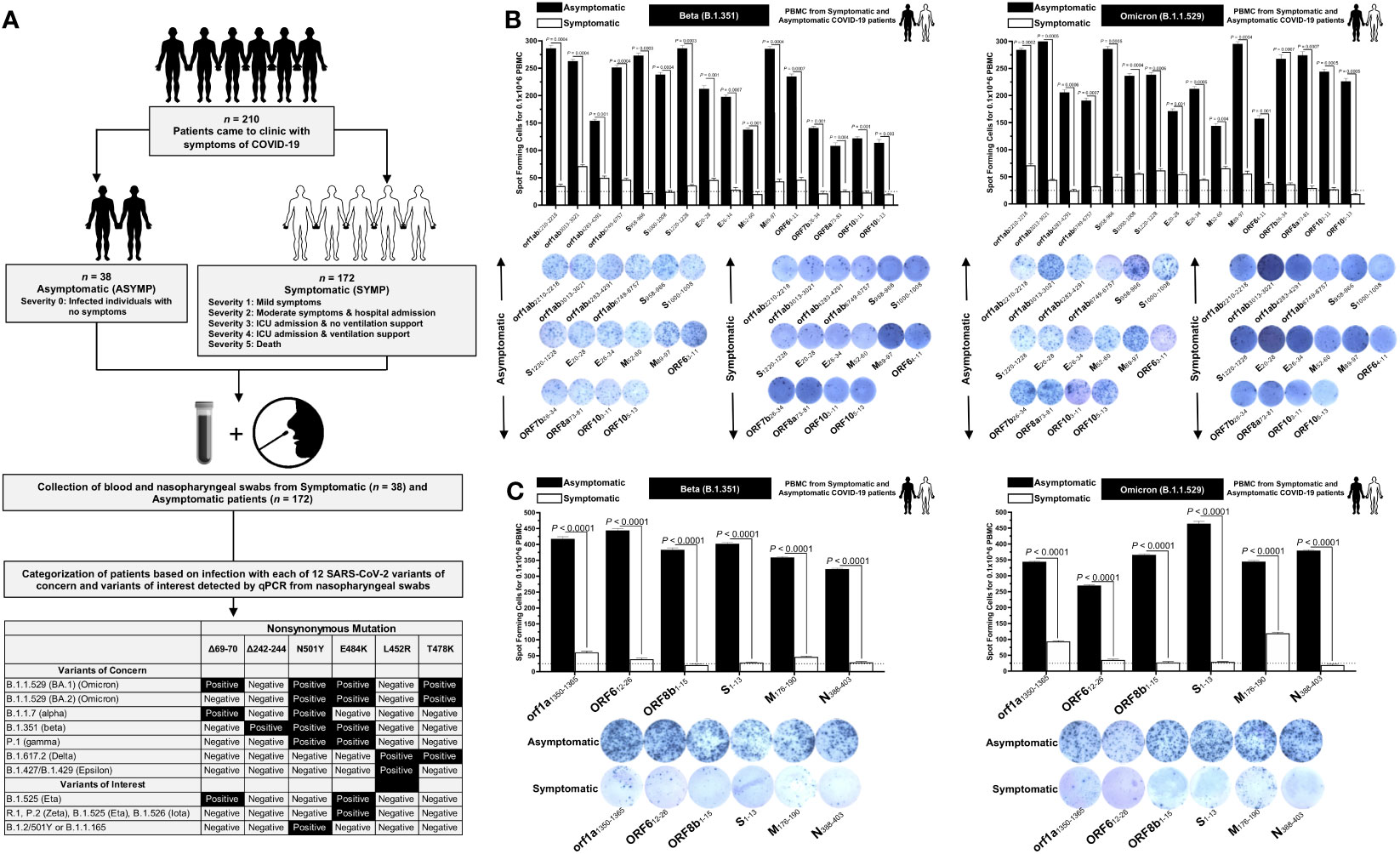
Frontiers Cross-protection induced by highly conserved human B, CD4+, and CD8+ T-cell epitopes-based vaccine against severe infection, disease, and death caused by multiple SARS-CoV-2 variants of concern
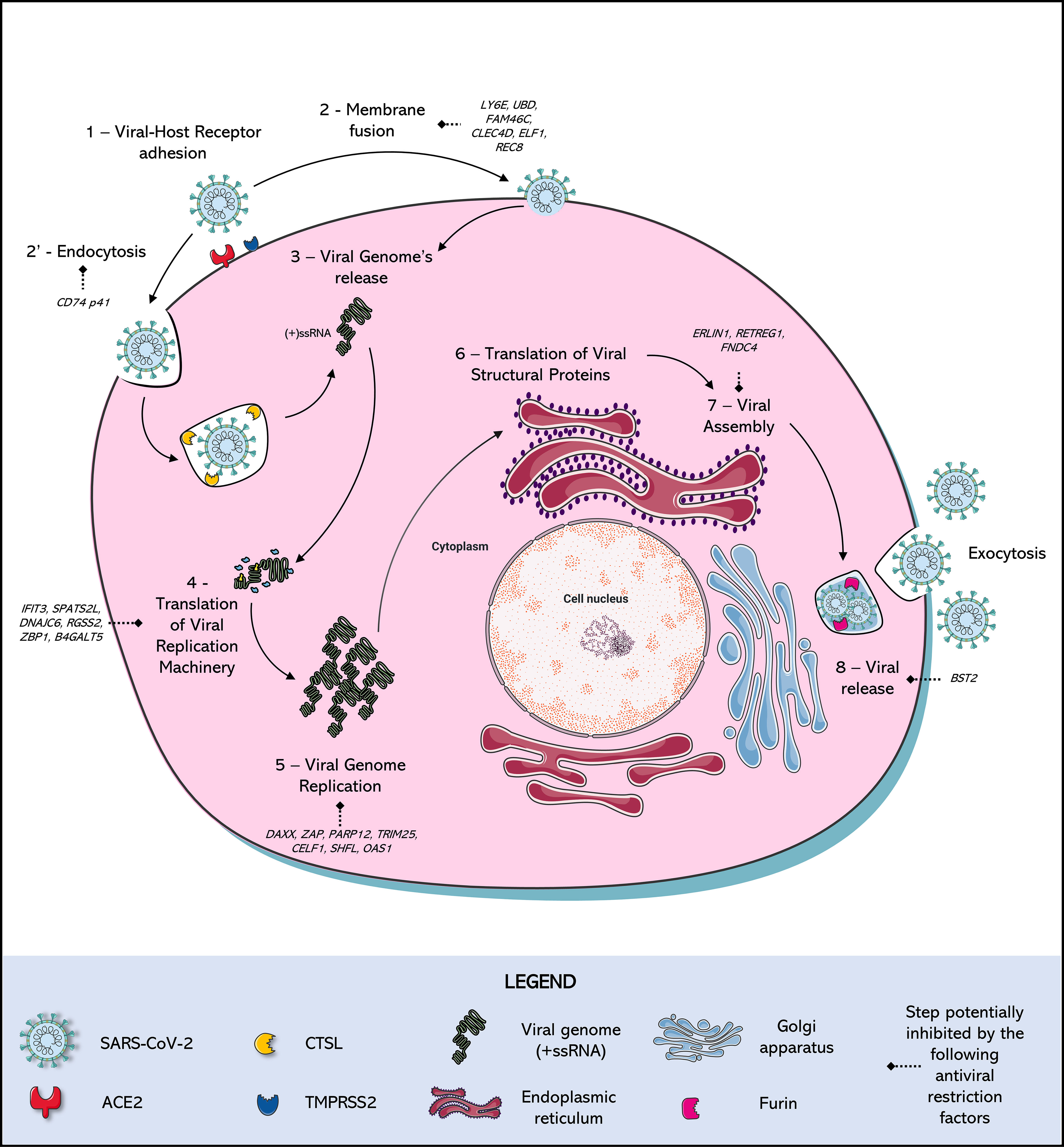
Frontiers Choosing a cellular model to study SARS-CoV-2

Overall structure of the RBD–EY6A complex a, A ribbon diagram of

Browse Preprints - Authorea

The race to understand immunopathology in COVID-19: Perspectives on the impact of quantitative approaches to understand within-host interactions - ScienceDirect

Identification and validation of 174 COVID-19 vaccine candidate epitopes reveals low performance of common epitope prediction tools
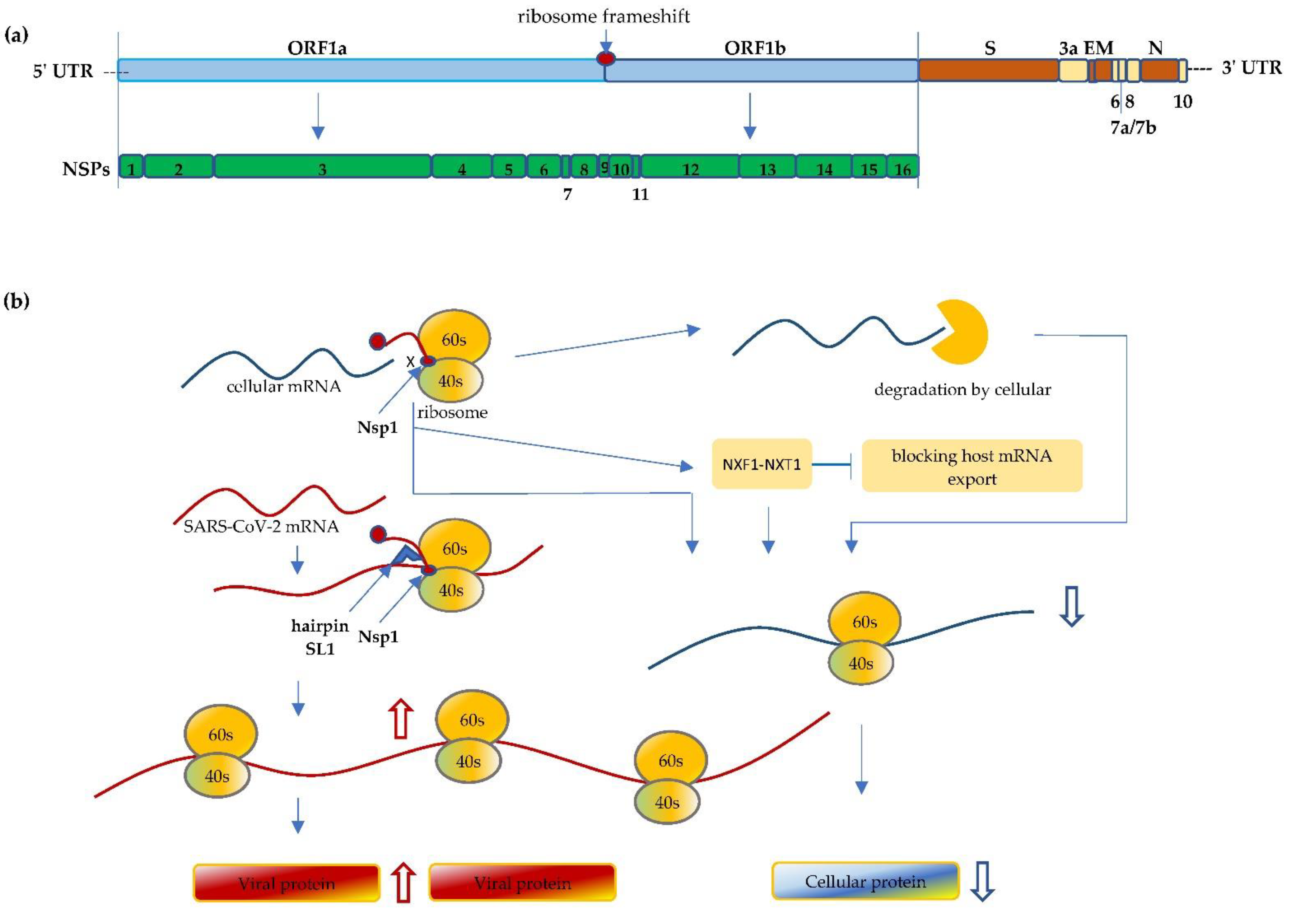
Life, Free Full-Text
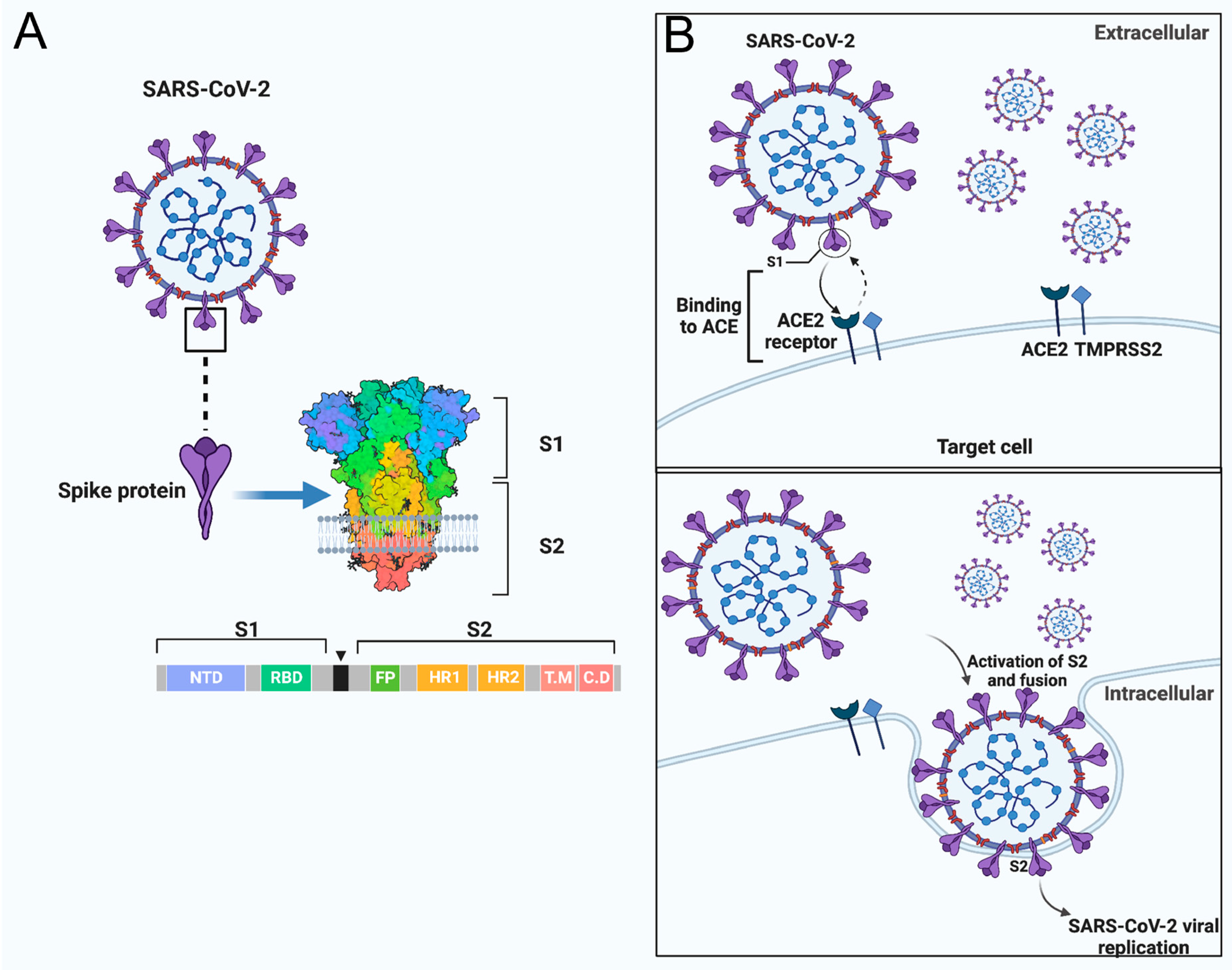
Vaccines, Free Full-Text

Identification and validation of 174 COVID-19 vaccine candidate

The precision and recall curve. The threshold estimations of

Structural basis for the neutralization of SARS-CoV-2 by an

EM structure of the SARS-CoV-2 spike–EY6A Fab complex a,b, Side (a

Mutational Analysis Of Circulating Omicron SARS-CoV-2

Evaluation of immune evasion in SARS-CoV-2 Delta and Omicron variants - Computational and Structural Biotechnology Journal
In silico prediction of immune-escaping hot spots for future COVID-19 vaccine design
- Naked Fiber Looks Good, Featured News Story

- 37 Motivational Exercise Quotes to Get You Through (or to) Your Next Workout
:max_bytes(150000):strip_icc()/01_MotivationalQuote_Gray-7c57fd7db73f49eea3623e8681963353.jpg)
- Women's Butt Lifting Yoga Shorts, Workout High Waist Tummy Control Ruched Booty Pants, Sexy Peach Butt Hip Lift Short

- Buy Mesh Corset Underbust Corsets for Women - Curvify Me

- Sassy Crop – Euphoria