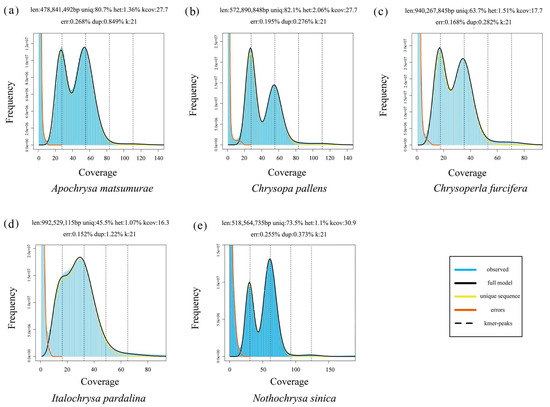
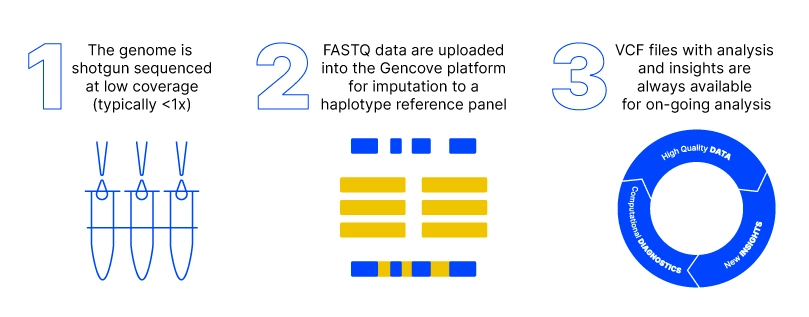
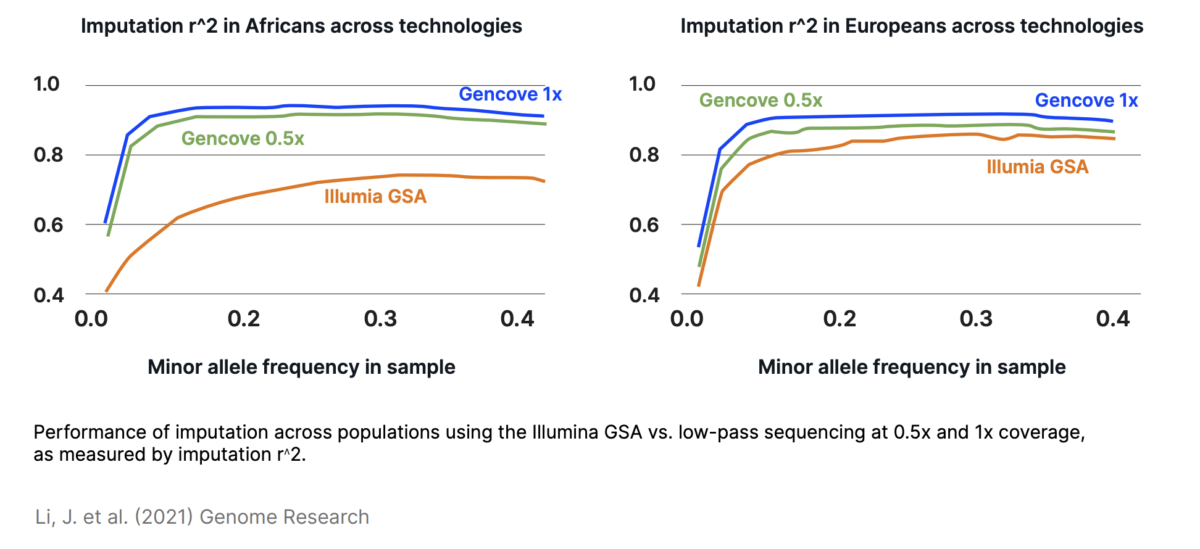
Low-pass sequencing and imputation for evaluating genetic
By A Mystery Man Writer

Best practices for analyzing imputed genotypes from low-pass sequencing in dogs

Genotype imputation within a sample of related individuals
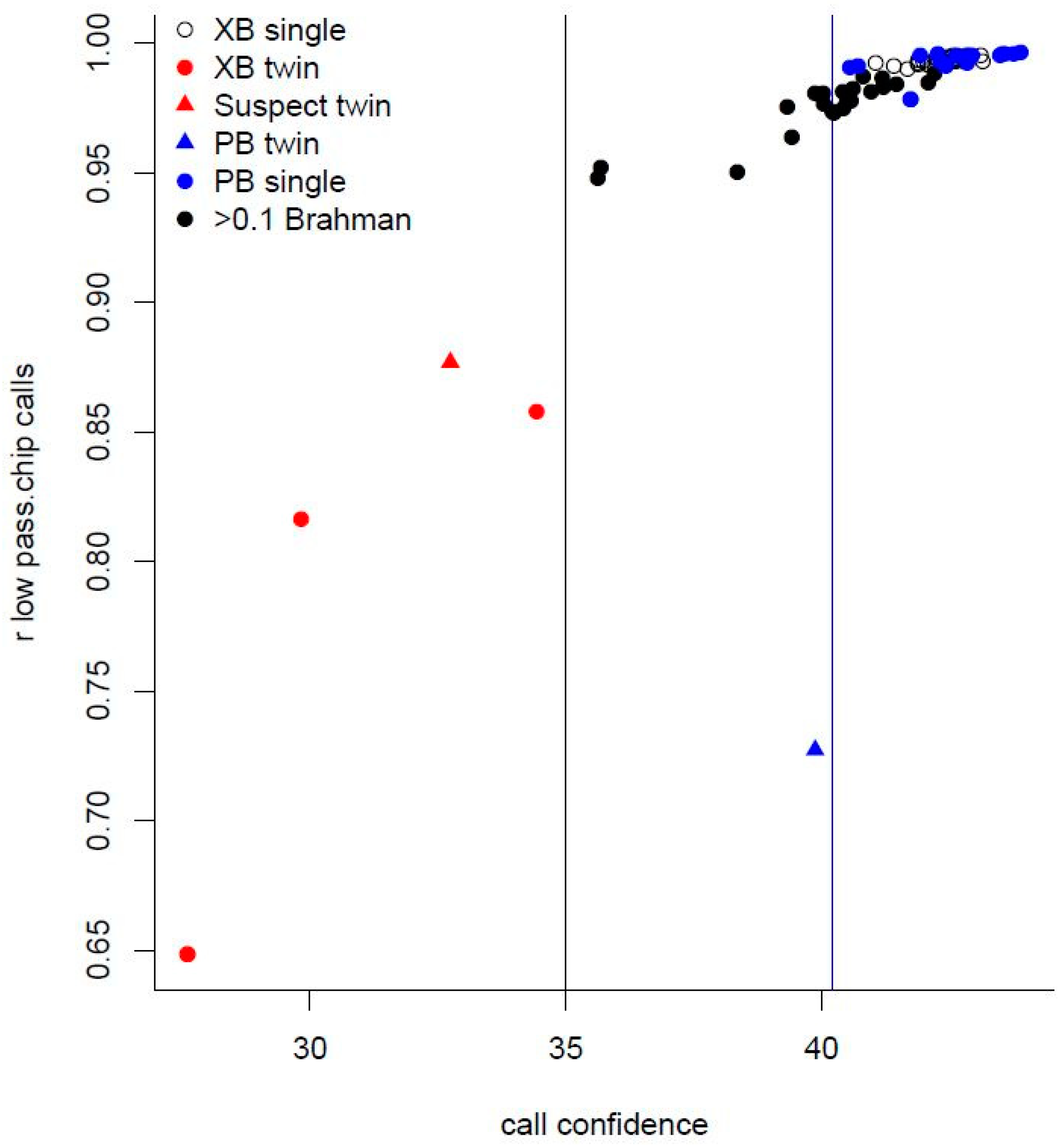
Genes, Free Full-Text

Proceedings of the 27th International Stroke Genetics Consortium Workshop

Low-Coverage Whole Genome Sequencing - NCI

Low-coverage whole genome sequencing for a highly selective cohort of severe COVID-19 patients
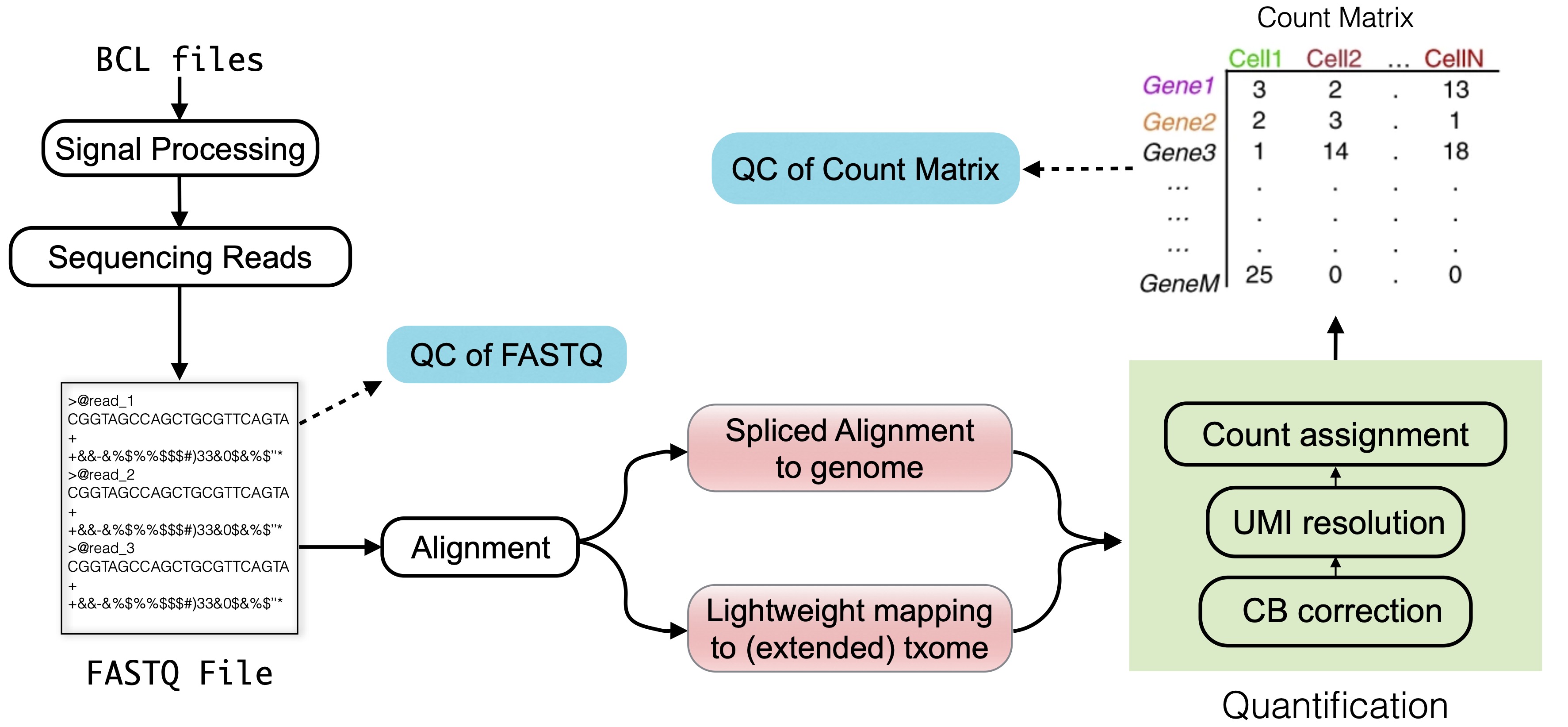
3. Raw data processing — Single-cell best practices
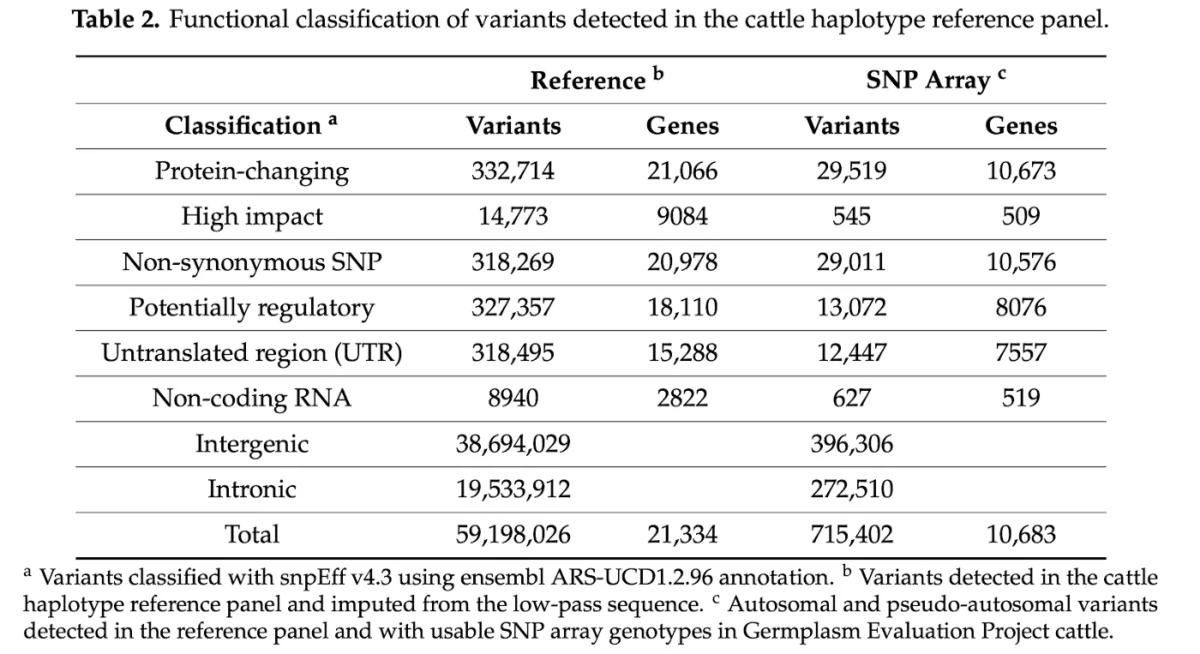
New publication in collaboration with the USDA: Assessment of Imputation from Low-Pass Sequencing to Predict Merit of Beef Steers - Gencove
Integrative genomics analysis identifies promising SNPs and genes implicated in tuberculosis risk based on multiple omics datasets

Evaluating the performance of dropout imputation and clustering methods for single-cell RNA sequencing data - ScienceDirect

PDF) Evaluating the concordance between sequencing, imputation and microarray genotype calls in the GAW18 data
Biobanks and Beyond: Genotyping for the Future

Low-coverage sequencing cost-effectively detects known and novel variation in underrepresented populations - ScienceDirect
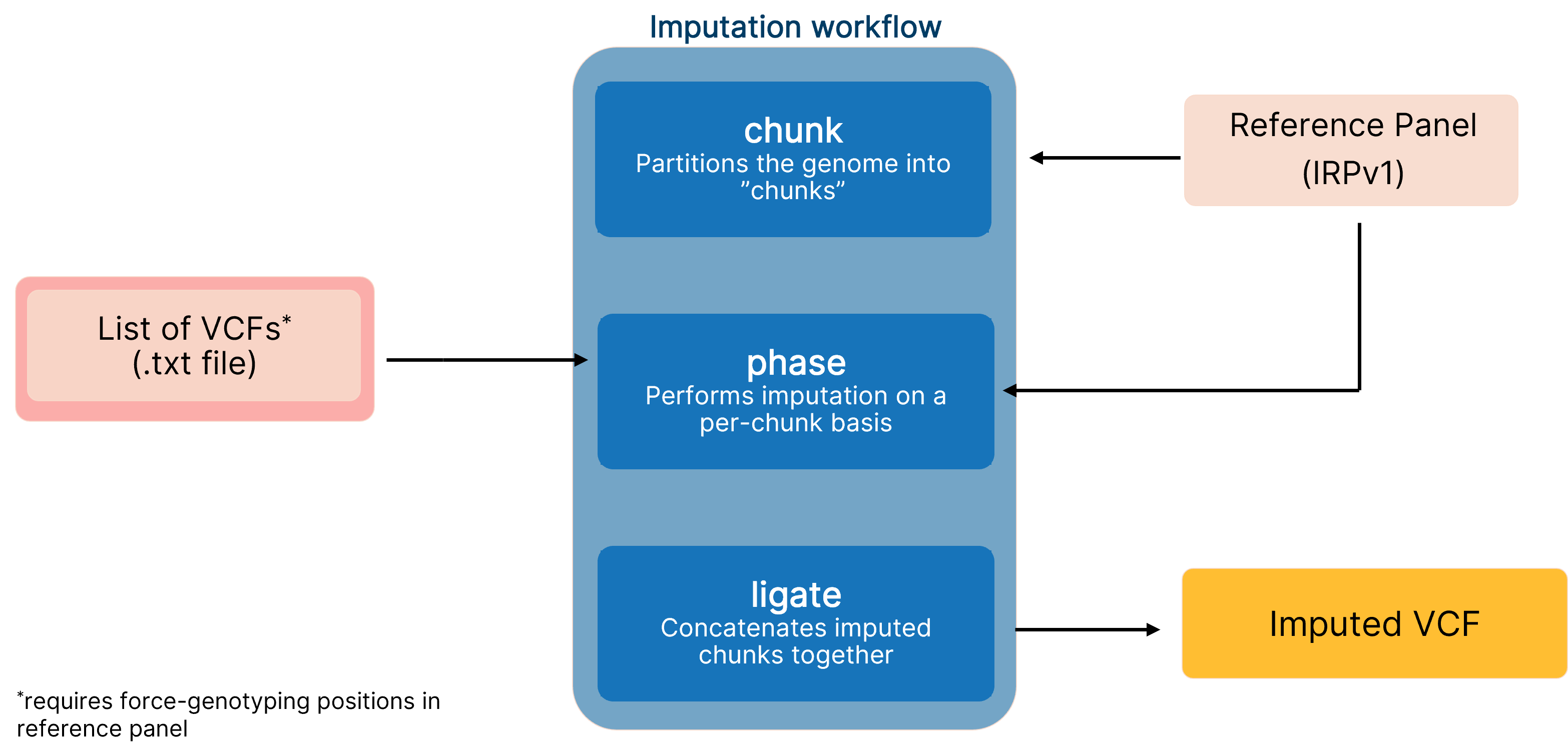
Boosting variant calling performance using a high-quality reference panel for imputing low-coverage sequencing data