Evaluating coverage bias in next-generation sequencing of Escherichia coli
By A Mystery Man Writer
Whole-genome sequencing is essential to many facets of infectious disease research. However, technical limitations such as bias in coverage and tagmentation, and difficulties characterising genomic regions with extreme GC content have created significant obstacles in its use. Illumina has claimed that the recently released DNA Prep library preparation kit, formerly known as Nextera Flex, overcomes some of these limitations. This study aimed to assess bias in coverage, tagmentation, GC content, average fragment size distribution, and de novo assembly quality using both the Nextera XT and DNA Prep kits from Illumina. When performing whole-genome sequencing on Escherichia coli and where coverage bias is the main concern, the DNA Prep kit may provide higher quality results; though de novo assembly quality, tagmentation bias and GC content related bias are unlikely to improve. Based on these results, laboratories with existing workflows based on Nextera XT would see minor benefits in transitioning to the DNA Prep kit if they were primarily studying organisms with neutral GC content.

CRISPR/Cas9 recombineering‐mediated deep mutational scanning of essential genes in Escherichia coli

Human genome - Wikipedia

Raw read quality control parameters. Raw sequence read QC parameters

Sequence bias informs k-mer bias during sequencing. Top

k-mer - Wikipedia
Evaluating coverage bias in next-generation sequencing of Escherichia coli

Multi-Platform Assessment of DNA Sequencing Performance using Human and Bacterial Reference Genomes in the ABRF Next-Generation Sequencing Study

PDF] Comparison of the sequencing bias of currently available library preparation kits for Illumina sequencing of bacterial genomes and metagenomes
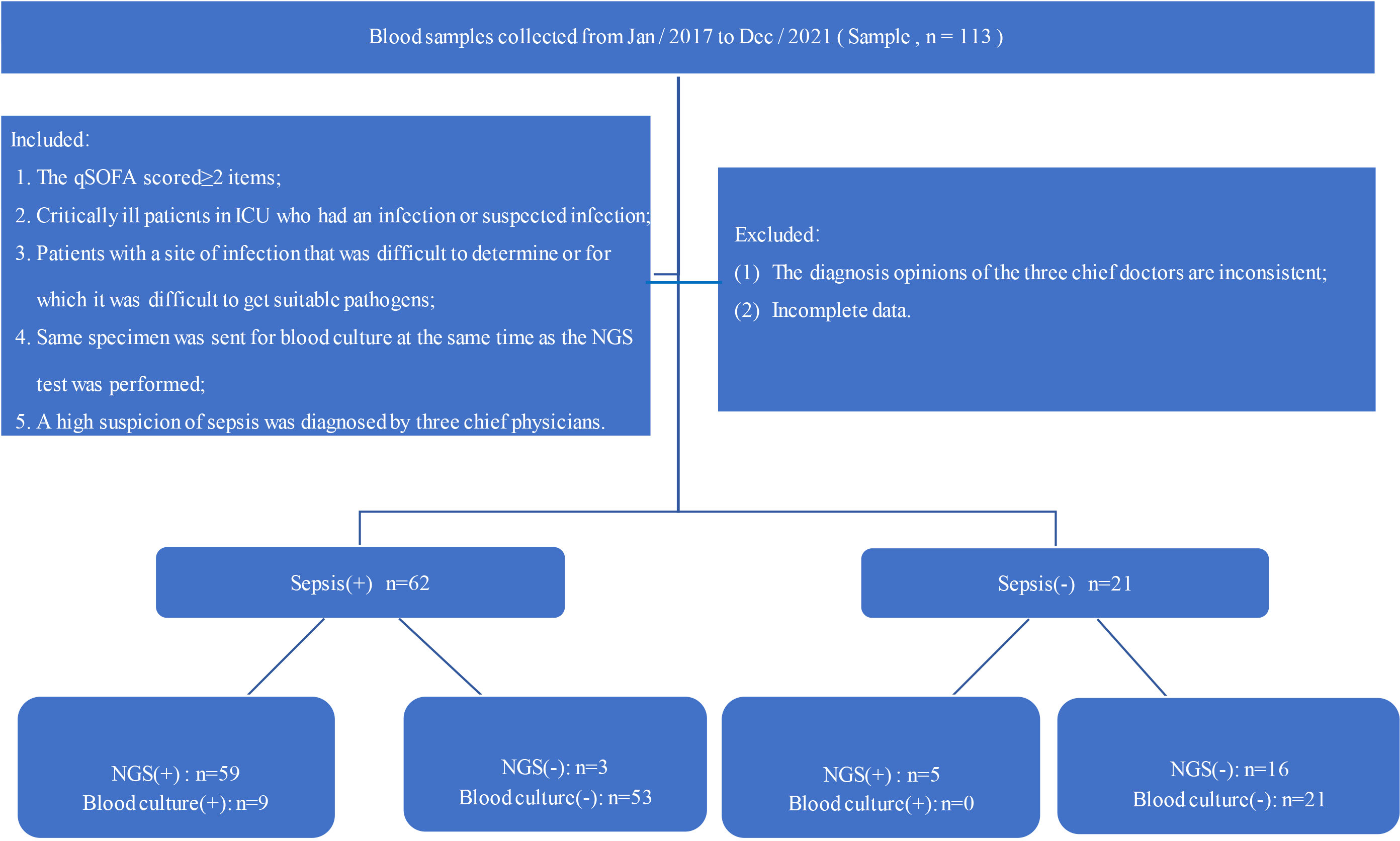
Frontiers The diagnostic value of next-generation sequencing technology in sepsis

Measuring sequencer size bias using REcount: a novel method for highly accurate Illumina sequencing-based quantification, Genome Biology
DNA sequencing - Wikipedia

Barplot showing the base counts (a) and boxplot showing the normalized

Sequencing biases of low-GC regions and assembly quality of entire

Coverage profiles of HRSV obtained per sample per methodology. A-I
Evaluating coverage bias in next-generation sequencing of Escherichia coli
- Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex
- Women Lace Briefs,Triangle Panties,Breathable Thin Briefs,Low Waist Thongs,Half Coverage Panties,Seamless Briefs,Women Night Underwear,5-Pack

- Vinyl truck lettering crew cab short bed low coverage premium

- Low-coverage sequencing cost-effectively detects known and novel

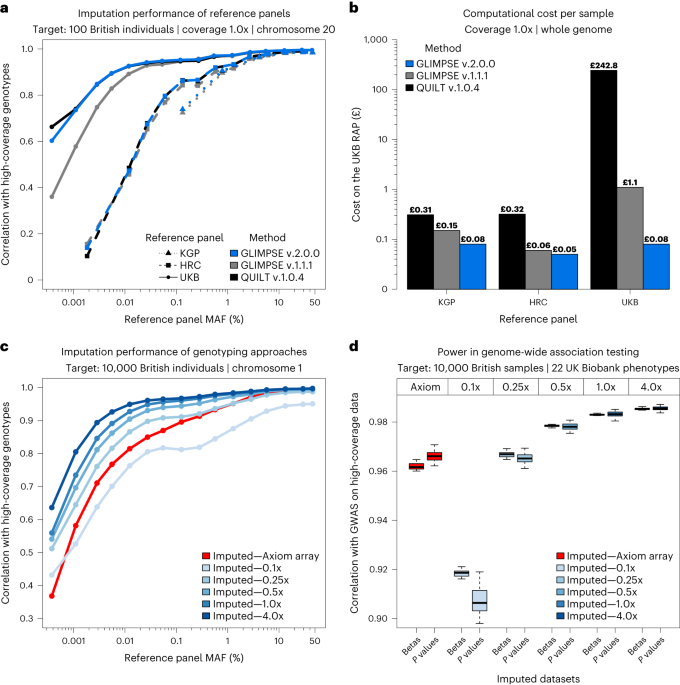
- Imputation of low-coverage sequencing data from 150,119 UK Biobank
- Shop POLO RALPH LAUREN 2022 SS Unisex Plain Logo Boxer Briefs

- Houndstooth fabric seamless pattern Royalty Free Vector

- Calm Club - The Oracle Shop Tarot Decks at Mom's - Mom's Jewelry

- 2023 New Upgrade High Waist Leak Proof Panties, Design Comfort

- Buy KHWAISH STORE Wirefree High Support Bra for Women Small to Plus Size Everyday Wear, Back Support, Non Padded Bra, Size(32 Till 50) Pack of 1 Black at
